

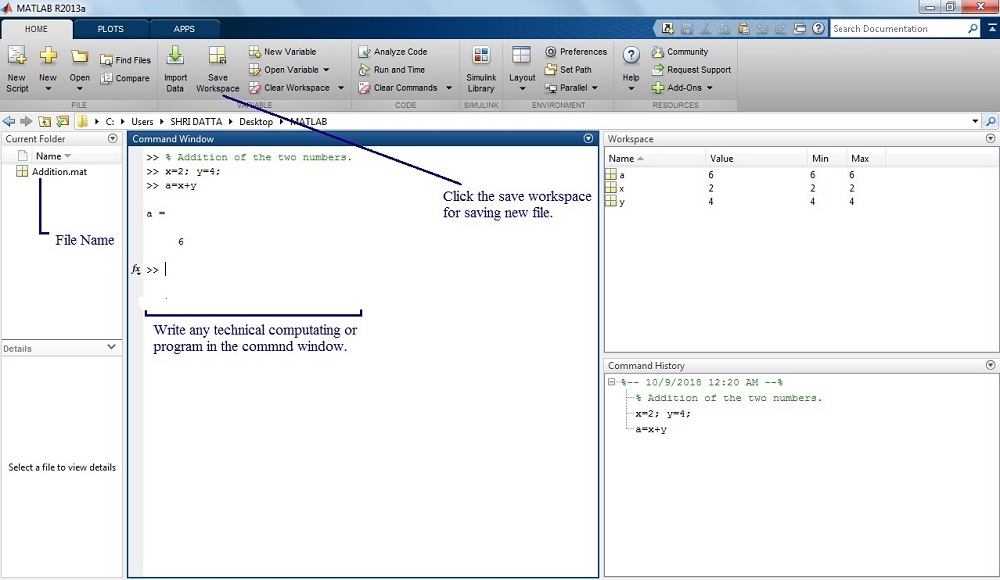
#perform RMA normalization (I would normally use GCRMA but it did not work with this chip) Raw.data=ReadAffy(verbose=TRUE, filenames=cels, cdfname="hugene10stv1") #From bioconductor Setwd("/Users/ogriffit/Dropbox/BioStars/GSE27447/data") Sapply(paste("data", cels, sep="/"), gunzip) Setwd("/Users/ogriffit/Dropbox/BioStars/GSE27447")Ĭels = list.files("data/", pattern = "CEL") #Download the CEL file package for this dataset (by GSE - Geo series id) Setwd("/Users/ogriffit/Dropbox/BioStars")
# install additional bioconductor libraries, if not already installedīiocLite("hugene10sttranscriptcluster.db") #install the core bioconductor packages, if not already installed
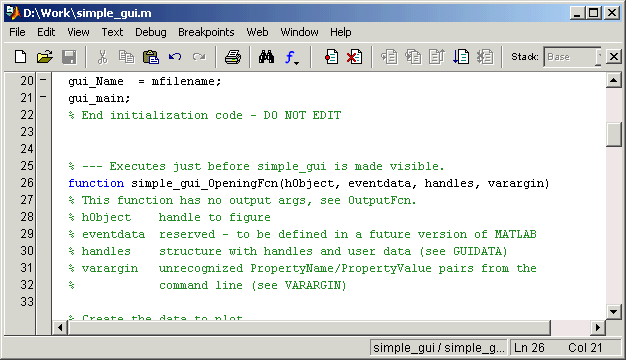
It assumes you have latest version of R installed and will have to change some working directories. This is how I would process that particular dataset in R/Bioconductor.


 0 kommentar(er)
0 kommentar(er)
